Illumina RNA Sequencing
RNA, the intermediate stage in the flow of genetic information from DNA to protein, contains many systems of expression and regulation that can be studied through RNA sequencing. While whole genome sequencing reveals information at the DNA level, next generation RNA sequencing (RNA-seq) characterizes the transcriptome and provides additional context to the genome with qualitative and quantitative expression profiles.
In contrast to the fixed state of the genome, the transcriptome’s variability in content and quantity provides a prime opportunity to study response mechanisms and control systems. For this reason, many RNA-seq projects compare transcript expression of an organism at different developmental stages, under various experimental conditions, and across multiple time points. Understanding the transcriptomic level changes between samples and conditions allows for the interpretation of functional regions of the genome and further extrapolation of how the resulting proteins function to support life.
The first step of any RNA-seq project is the isolation of total RNA from the sample of interest. Total RNA is composed of (among other things) both ribosomal RNA (rRNA) and messenger RNA (mRNA). While rRNA constitutes as much as 90% of total RNA, it is ubiquitous and often provides little in the way of helpful signals. Conversely, mRNA typically makes up only 1-2% of the sample but contains all of the expression information. Successful and cost-effective transcriptome profiling and multiplexing relies on focused sequencing of transcripts of interest, made possible through the removal of unwanted ribosomal material. Thus, a defining factor in any RNA-seq study choosing between one of two approaches to narrow in on the target content: rRNA depletion (negative selection) and polyA enrichment (positive selection). For additional information about the two strategies and pricing, please visit the individual service pages below.
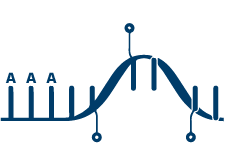
mRNA Enrichment
PolyA enrichment positively selects for transcripts with polyadenylated tails and is often referred to as mRNA sequencing. This method provides a clean way of sequencing only on-target fragments but is only applicable for organisms that create polyA-tailed transcripts. Because of this, polyA enrichment sequencing is generally the choice for eukaryote-only studies.
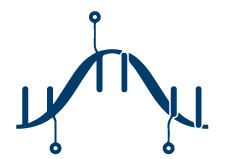
rRNA Depletion
rRNA depletion enzymatically degrades the ribosomal RNA fragments from a sample by relying on probes targeted at rRNA sequences specific to the organism. It is often (and confusingly) referred to as total RNA sequencing. rRNA depletion works well for prokaryotes and will capture all RNA molecules over 150 bp that are not degraded.
While these guiderails broadly apply, there is a nuance to selecting the most effective sequencing strategy for each project, with differing advantages and disadvantages to each method and biases that impact downstream analyses. We recommend visiting both of our specialty pages for more detailed descriptions to explore the options. If you are still unsure which method is best for your work, please do not hesitate to contact us.
RNA Workflow Overview
For both methods, SeqCenter uses Illumina library prep kits which include DNase treatment as the first step. This will remove any residual genomic content, but primary DNase treatment immediately after harvesting is recommended for maximum removal. To minimize degradation, samples continue straight through to the targeting method of choice, cDNA synthesis, and library preparation. Final libraries are run on high-throughput and accurate Illumina NovaSeq platforms. The resulting 2x150bp sequencing reads are distributed as compressed fastq files. Additionally, each order will receive a project report with all methods specific to your samples for ease of publication at a later date. We offer Basic and Intermediate Data Analysis packages for those seeking additional bioinformatics assistance.
Maximizing the Power of Your RNA Experiment
In order to obtain high-quality and biologically meaningful data from an RNA-seq experiment, careful experimental design is critical. In particular, attention must be paid to details such as sample preparation and storage, the level of biological replication, sequencing read depth, and batch effects.
Additional Material Considerations for All RNA Submissions
- Material requirements for each service ensure library preparation success and high numbers of unique reads. Because the target material makes up a very tiny fraction of the total RNA, samples under 1 µg total have very limited chances of successful library preparation and could result in low read diversity. Successful preps for low concentration samples tend to indicate unsuccessful depletion and result in mostly ribosomal sequences.
- Library preparation must continue to move from total RNA to cDNA without pausing due to the fragile nature of RNA. Because of this, there are not any holding points during library prep, and we are not able to assess sample quality without seeing the entire prep through.
- Samples that are below the minimum requirements for success after the first step of DNase treatment will be removed from prep and will not be processed. All other samples that pass this threshold from the order will move forward. The customer will be notified of any failures at the time of distribution.
- If you know in advance that any sample failures affect whether or not other samples should be sequenced (because of batch effect or loss of analytical power), please contact us to discuss these arrangements before submission.
- If you know in advance that you would like SeqCenter to attempt library preparation for any samples below the 1µg total threshold, please make these arrangements before submission. Samples below the threshold that are processed will be billed for the full price of the sequencing package, regardless of sequencing output.
- RNA integrity directly correlates to depletion efficiency, due to the targeted nature of both approaches. SeqCenter strongly recommends that customers assess fragment length before submission and does not perform fragment analysis as part of our standard pipelines. Additionally, RIN calculations can give some estimation of degradation but can vary greatly. Because of this, SeqCenter gives only an optional recommendation of RIN > 6.
Stranded Library Preparation
All RNA libraries that SeqCenter generates capture the original transcripts’ strand information. This data is critical in the identification of overlapping genes, appropriate splicing, and for aligning reads to poorly annotated reference genomes. In addition, many bioinformatic analyses require this information to ease the alignment process, and studies show that stranded information provides a more accurate estimate of transcript abundance.
SeqCenter relies on Illumina’s stranded technology to translate strand-specificity through library preparation and sequencing. Briefly, stranded information is captured through the use of dUTPs (instead of dTTPs) in the second strand synthesis step of cDNA synthesis. After adaptor ligation, second strand amplification will be suppressed in the final library amplification due to polymerase stalling at the location of the incorporated dUTPs. Due to directionality of sequencing adaptors, Read 1 (R1) will always map to the antisense strand and Read 2 (R2) will always map to the sense strand. How this process works in more detail can be found at Illumina’s knowledge link.
If you prefer to have a non stranded library preparation, please contact us with a description of your project to discuss what services we can provide.
Note for Small RNA Projects
We do not offer a standard library preparation service to capture microRNAs (miRNA) or small RNAs less than 150bp in length. If you are looking to sequence micro-RNAs or RNAs smaller than 150bp, please contact us to discuss custom services we can provide for your project.
Additional Resources:
- Hansen, K. D., Wu, Z., Irizarry, R. A. & Leek, J. T. Sequencing technology does not eliminate biological variability. Biotechnol.29, 572–573 (2011). Required reading for anyone considering RNA-seq or other -omics technologies. A well-written reminder of why quantitative RNA experiments will always need replicates, even if RNA assay technologies were perfect. The authors caution users against being overenthusiastic about new technologies and discarding lessons learned about experimental design.
- Wu, H., Wang, C. & Wu, Z. PROPER: comprehensive power evaluation for differential expression using RNA-seq. Bioinformatics.31, 233–241 (2015).
- Gaye, A. Extending the R Library PROPER to enable power calculations for isoform-level analysis with EBSeq. Genet.7, 225 (2017).
- The following are model-based examinations of minimum reads required for some model organisms. It is important to note that these are for minimal statistical power and bare minimum numbers to achieve it. Buffering room should be allowed for less than 100% depletion efficiency and noisy data.
- Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ, Livny J, Earl AM, Gevers D, Ward DV, Nusbaum C, Birren BW, Gnirke A. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 2012;13(3):R23. doi: 10.1186/gb-2012-13-3-r23. PMID: 22455878; PMCID: PMC3439974.
- Haas, B.J., Chin, M., Nusbaum, C. et al. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes?. BMC Genomics 13, 734 (2012).
- Schurch N. J., Schofield P., Gierlinski M., Cole C., Sherstnev A., Singh V., et al. (2016). How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA N. Y. N 22, 839–851. 10.1261/rna.053959.115
- Lamarre S, Frasse P, Zouine M, Labourdette D, Sainderichin E, Hu G, Le Berre-Anton V, Bouzayen M, Maza E. (2018). Optimization of an RNA-Seq Differential Gene Expression Analysis Depending on Biological Replicate Number and Library Size. Front Plant Sci. 2018 Feb 14;9:108. doi: 10.3389/fpls.2018.00108
- Liu Y., Zhou J., White K. P. (2014). RNA-seq differential expression studies: more sequence or more replication? Oxf. Engl. 30, 301–304. 10.1093/bioinformatics/btt688
- Ching T., Huang S., Garmire L. X. (2014). Power analysis and sample size estimation for RNA-Seq differential expression. RNA20, 1684–1696. 10.1261/rna.046011.114
- Palazzo, Alex & Lee, Eliza. (2015). Non-coding RNA: What is functional and what is junk?. Frontiers in genetics. 6. 2. 10.3389/fgene.2015.00002.
- Levin, J. Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–715 (2010).

Contact:
91 43rd Street, Ste. 250
Pittsburgh, PA 15201
(878) 227-4915
Services:
About:
Resources:
